Illumina read filtering - without merging - HPC guide
In this tutorial we will process raw Illumina reads for downstream mapping/assembly. Download and extract the following read 1 and read 2 data files.
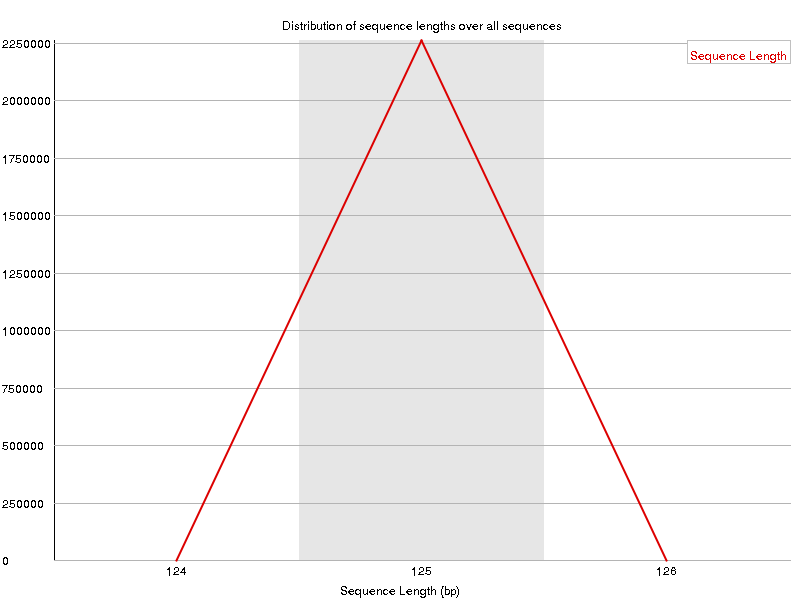
Reads are 125 bp long from a size selection of ~300-550 bp (tapestation result). We expect that none of these reads will overlap and, therefore, skip merging.
Create a project directory, sub-folders and import data
‘mkdir project’
‘cd project’
‘mkdir 00-raw 01-fastqc 02-remove_contaminants 03-trim_reads 04-demultiplex’
Move the raw Illumina reads (.fastq.gz format) to the 00-raw directory within the new project
I use the FTP client FileZilla to transfer between PC/HPC.
1. Quality checking using Fastqc
Navigate to the 01-fastqc folder
‘cd 01-fastqc’
This script will run fastqc - email is optional, remove last two #SBATCH lines if needed
‘nano run_fastqc’
Paste in the following italicised text;
#!/bin/bash
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=4
#SBATCH --mem=4G
#SBATCH --time=00-02
#SBATCH --mail-user=name@place.com
#SBATCH --mail-type=ALL
module load fastqc
fastqc ../00-raw/*fastq.gz -o ./
‘Ctrl x’ to exit and ‘y’ to save
Now we can submit the job by typing ‘sbatch run_fastqc’
Once fastqc is complete type ‘ls-1’ and you will see a .html and a .zip (not shown) for each raw file
[user@hpc 01-fastqc]$ ls -1
IDX11_CD25JANXX_GGCTAC_L006_R1_fastqc.html
IDX11_CD25JANXX_GGCTAC_L006_R2_fastqc.html
IDX12_CD25JANXX_CTTGTA_L006_R1_fastqc.html
IDX12_CD25JANXX_CTTGTA_L006_R2_fastqc.html
Transfer the .html files to your PC and open it in any internet browser
The quality of this data is good, although it is typical of ‘low diversity’ RADseq data
2. Trim reads
Now we will process our reads by using three filtering steps in the program Trimmomatic. First, we will filter for quality. We do this by sliding along the read and averaging the quality across 4 bases, the end of the read will be trimmed off once the phred score drops below 20 (99% base call accuracy- set by SLIDINGWINDOW:4:20). Now that reads have been trimmed for quality, reads <100 bp are removed (MINLEN:100). Finally, we will filter out Illumina adapters (ILLUMINACLIP:TruSeq3-PE-2.fa:2:30:10:1:true) from our reads by providing sequences in a file within the 02-trim_reads directory.
Download: TruSeq3-PE-2.fa
Note: the order of trimming steps can be changed by swapping the order that each of the above three processes are listed in the script.
Navigate to the trimming directory ‘cd ../02-trim_reads/’
‘nano trim_reads’
Paste in the following italicised text;
#!/bin/bash
#SBATCH --nodes=1
#SBATCH --ntasks=2
#SBATCH --cpus-per-task=4
#SBATCH --mem=32G
#SBATCH --time=00-01
echo 'Starting at:' `date`
module load Trimmomatic parallel/20170206-GCC-6.2.0 fastqc
for i in 11 12; do
echo "java -jar $EBROOTTRIMMOMATIC/trimmomatic-0.38.jar PE \
../00-raw/IDX"$i"*_R1.fastq.gz \
../00-raw/IDX"$i"*_R2.fastq.gz \
./IDX"$i"_R1_trim.fastq.gz \
./IDX"$i"_R1_unpaired_trim.fastq.gz \
./IDX"$i"_R2_trim.fastq.gz \
./IDX"$i"_R2_unpaired_trim.fastq.gz \
SLIDINGWINDOW:4:20 \
MINLEN:100 \
ILLUMINACLIP:TruSeq3-PE-2.fa:2:30:10:1:true LEADING:3 TRAILING:3 \
-threads 4"; done | parallel -j 2 --delay 1
fastqc ./*fastq.gz -o ./
echo 'Finished at:' `date`
‘Ctrl x’ to exit and ‘y’ to save
Now we can submit the job by typing ‘sbatch trim_reads’
Transfer the ‘IDX11_R1_trim_fastqc.html’ file to your PC and open it in any internet browser. You can see that the read length distribution has changed as we have removed the ends of the reads with quality issues. You can also look at the other three .html files. Our quality looks great and we haven’t lost too many reads by setting our minimum length to 100 bp. These can be adjusted if needed.
Type “more slurm-*.out” to investigate how many reads were retained. If you need to increase the percentage of reads retained you can adjust the phred and minimum length values based on the fastqc report.
E.g. Input Read Pairs: 2927828 Both Surviving: 2510040 (85.73%) Forward Only Surviving: 215216 (7.35%) Reverse Only Surviving: 96676 (3.30%) Dropped: 105896 (3.62%).
3. Remove contaminants - i.e. bacteria, archaea, and virus reads
Now we will filter out microbial contamination using Kraken. This requires a database to be installed on your HPC. Contact your admin person to for assistance with installing software and databases. We are using the ‘standard database’ from the Kraken link above. This is large and requires a lot of memory to run. In our script below we will select a specific partition with access to enough memory and allocate more RAM and CPUs.
For fun we will use the program Krona to produce an interactive pie chart that will help us visualise the taxonomy of our microbial hits.
Navigate to the filtering folder ‘cd ../03-remove_contaminants’.
‘nano release_kraken’
Paste in the following italicised text;
#!/bin/bash
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --partition bigmem
#SBATCH --cpus-per-task=8
#SBATCH --mem=256G
#SBATCH --time=00-05
#SBATCH --mail-user=name@place.com
#SBATCH --mail-type=ALL
echo 'Starting at:' `date`
module load Kraken Krona/2.7 parallel/20170206-GCC-6.2.0
database=standard
standard_db=/data/projects/KRAKEN/standarddb
for i in IDX11_R1_trim \
IDX11_R2_trim \
IDX12_R1_trim \
IIDX12_R2_trim
do
kraken ../02-trim_reads/"$i".fastq.gz --db $standard_db/ \
--unclassified-out "$i"_unc.fastq \
--classified-out "$i"_classified_$database.fastq \
--fastq-input --gzip-compressed \
--output "$i"_output_$database \
--threads 8; done
ktImportTaxonomy -t 3 -s 4 -c -o kraken_classified_microbe.html ./*_output_*
#-c combines data from all Kraken output files into a single Krona chart
#-t 3 -s 4 sets the correct columns for Krona to read in our Kraken output files.
gzip *_output_*
echo 'Finished at:' `date`
You will need to change the ‘standard_db=’ path to your Kraken database directory.
‘Ctrl x’ to exit and ‘y’ to save
The resulting files are labelled as follows:
Unclassified reads - used for downstream analyses
IDX11_R1_trim_unc.fastq
IDX11_R2_trim_unc.fastq
IDX12_R1_trim_unc.fastq
IDX12_R2_trim_unc.fastq
Interactive pie chart showing taxonomy of classified reads
kraken_classified_microbe.html
The slurm file acts as a log file when running Kraken. Look at it by typing ‘more slurm-*.out’
Starting at: Thu Feb 14 10:07:02 AEDT 2019
…
166571 sequences classified (8.70%)
1747292 sequences unclassified (91.30%)
…
Finished at: Thu Feb 14 10:09:25 AEDT 2019
The percentages (bold) show we have retained >90% of our reads. <9% of our first read file were matches for the microbial database. By opening the .html file in a web browser we can look at the taxonomy of our classified reads.
4. Demultiplex - based on barcodes
Now we can use Stacks to allocate our filtered non-merged reads to individuals based on the barcoded adapters added during library preparation. I use the adapters from Peterson et al (2012).
Navigate to the demultiplexing directory ‘cd ../04-demultiplex/’
You will need to add a file containing the barcodes used in your project to the ‘04-demultiplex’ directory. I have provided a file with the compete list of barcodes here. You can use the complete set, or create a file containing a subset.
Note: process_radtags will name your files according to their sample name if you add a second column to the barcode file. Example:
aagga site1_sample1
ggttg site1_sample2
caacc site2_sample1
…
‘nano run_process_radtags’
Paste in the following italicised text;
#!/bin/bash
#SBATCH --nodes=1
#SBATCH --ntasks=2
#SBATCH --cpus-per-task=8
#SBATCH --mem=32G
#SBATCH --time=03-00
echo 'Starting at:' `date`
module load Stacks parallel/20170206-GCC-6.2.0
barcode_file=./barcodes
enzyme=ecoRI
for i in 11 12; do
echo "mkdir IDX"$i"_demultiplexed && process_radtags -P " \
"-1 ../03-remove_contaminants/IDX"$i"_R1_trim_unc.fastq " \
"-2 ../03-remove_contaminants/IDX"$i"_R2_trim_unc.fastq " \
"-o ./IDX"$i"_demultiplexed " \
"-b $barcode_file -e $enzyme -i gzfastq " \
"&& gzip ./IDX"$i"_demultiplexed/*.fastq";
done | parallel -j 2 --delay 1
echo 'Finished at:' `date`
Note: this script specifies that reads are paired end reads (-P) and points to read 1 and read 2 for each index.
(-1 ../03-remove_contaminants/IDX"$i"_R1_trim_unc.fastq)
(-2 ../03-remove_contaminants/IDX"$i"_R2_trim_unc.fastq)
Note: downstream assembly packages such as ipyrad and Stacks accept independent paired end read 1 and read 2 files. Just be sure to specify this before assembly.